An Overview of COVID-19 Testing
Step 1: COVID-19 RNA Extraction
RNA extraction is the process by which RNA, the genetic material of the SARS-CoV-2 virus, is isolated and purified from a biological sample (1). In most cases, the biological sample used for COVID-19 testing is a nasal or oral swab. There are several ways to prepare biological samples for diagnostic testing. At Oak Crest, our sample preparation phase is known as a spin column-based nucleic acid purification (2). This method uses special filter membranes, usually made of derivatized silica or glass fiber, which are positioned at the bottom of a small plastic cylinder. These filters bind nucleic acids (i.e., RNA and DNA), but allow extraneous material to pass through during the purification process.
Before a sample can be tested for the presence of the virus, it must undergo a process of RNA isolation (3). To begin, the sample is treated with a lysis buffer, which is a solution that chemically breaks up cells and proteins within the sample to release nucleic acids from human, bacterial, and viral sources (4). In addition to chemical lysis, a brief vortex is performed to mechanically aid in the breakup of the cells and proteins in the sample. Next, the samples are placed in a centrifuge and spun at a precise speed, causing cellular debris and the lysis buffer to pass through the column and into a collection tube, leaving the RNA bound to the filter.
Subsequently, the samples go through several wash cycles with special buffer solutions to remove any residual impurities from the filter. Samples are centrifuged following each new addition of wash buffer solution, and the flow-through material is discarded. Finally, the column is placed in a new collection tube, and a small volume of highly purified water is added to the column. During centrifugation, the water causes the nucleic acids to un-bind from the filter and flow through the column into the new tube. This step is known as elution and marks the end of the RNA extraction protocol (5).

The COVID-19 testing team at Oak Crest Institute of Science processes samples three times per week to maintain a safe workspace.
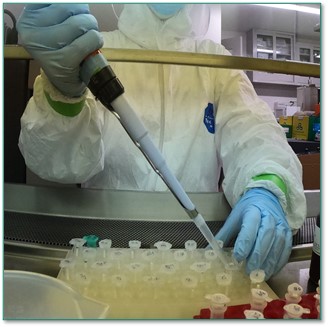
Lysis buffer is added to each swab to break up the samples.
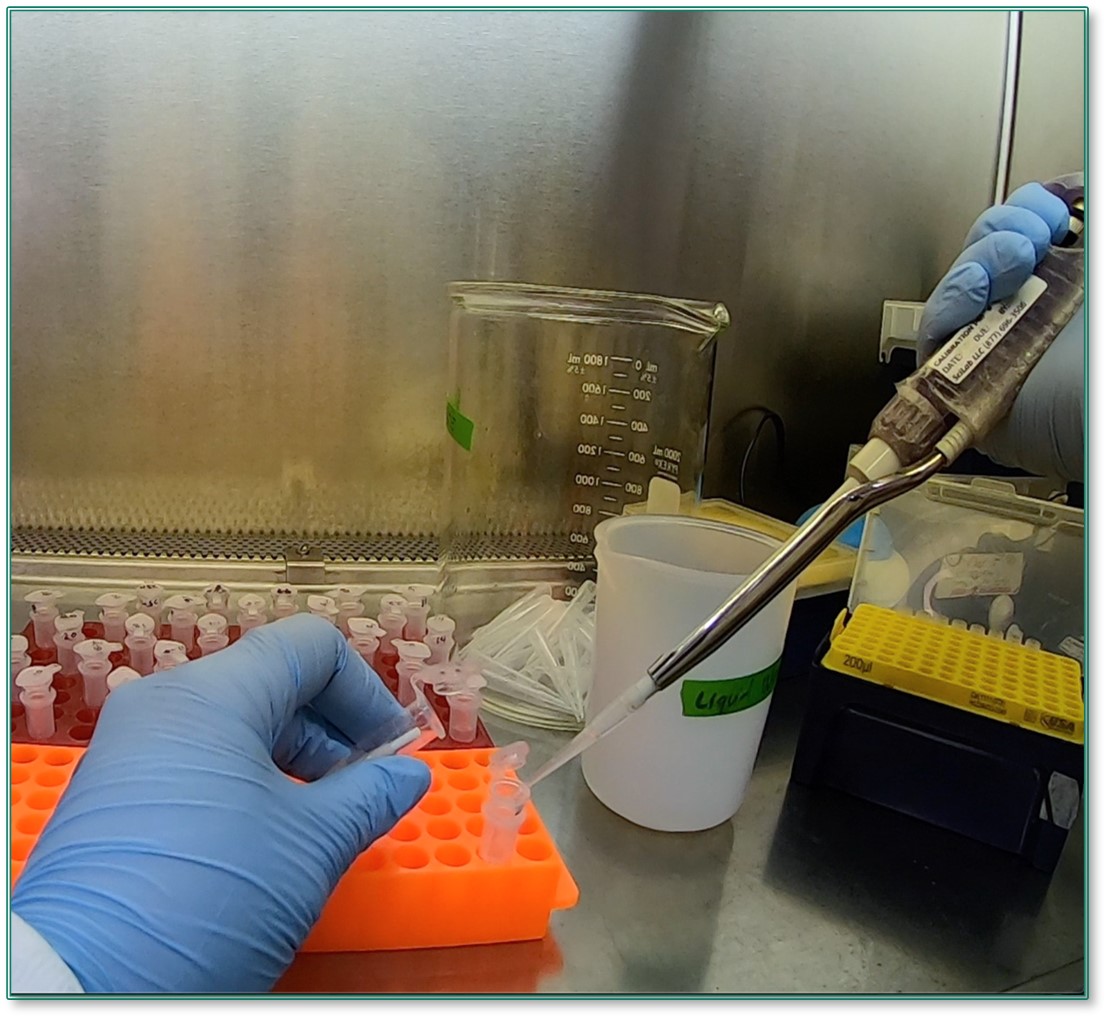
After adding lysis buffer and vortexing, the samples are transferred to spin columns to undergo RNA isolation.
Step 2: COVID-19 RT-qPCR
After extracting and purifying the RNA, it is time to perform a reverse transcription quantitative polymerase chain reaction (RT-qPCR) assay (6). By this point, however, each purified sample contains massive amounts of genetic material from various sources, such as human and bacterial RNA, as well as potential genetic information from other unrelated viruses. How do we identify the presence of a single gene sequence in a relative ocean of genetic material present in each purified sample? The answer is specific and exponential amplification of the gene of interest (6).
The qPCR technique works on the above principle. Initially, all the RNA in the sample is converted to DNA. Then, qPCR makes use of fluorescent probes, which are short DNA templates that are designed to bind to a specific region of the DNA segment that is being amplified (7). In this assay, the probes are specific to the SARS-CoV-2 virus and fluoresce only when the viral gene sequence of interest is present. Finally, the PCR reaction enables the detection and quantitation of the fluorescent signal for each cycle of amplification it runs. This means that the increase in intensity of the fluorescence is directly proportional to the increasing quantities of the target DNA sequence in a positive sample (8). These sequence-specific fluorescent probes are just one component in the overall qPCR cocktail of reagents and are packaged in a special master-mix reagent (9). The mix also contains reverse transcriptase, an enzyme that is necessary for creating complementary DNA (cDNA) from an RNA template (10). Because the genetic material of SARS-CoV-2 is single-stranded RNA, the gene sequences first must be converted to double-stranded DNA before the process of gene amplification can begin. Additionally, the mix contains DNA polymerase and dNTPs (building blocks of DNA), which are necessary to produce more copies of DNA as the target sequence is amplified (11).
Primers, short single-stranded pieces of DNA that attach to specific locations on the gene of interest, are added separately from one another to the rest of the qPCR components (12). Each primer is tested separately by qPCR to allow for greater diagnostic accuracy by testing the presence of more than one viral gene sequence. For diagnostic testing of SARS-CoV-2, two primer pairs are included, both of which coincide with different portions of the viral genome that encode a protein unique to the virus (13). Additionally, a primer for a specific human gene sequence is also included to ensure sufficient sample collection (14).
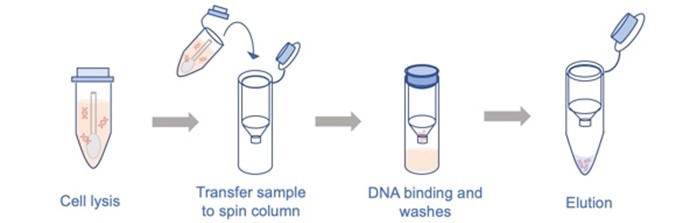
Schematic of RNA extraction process.
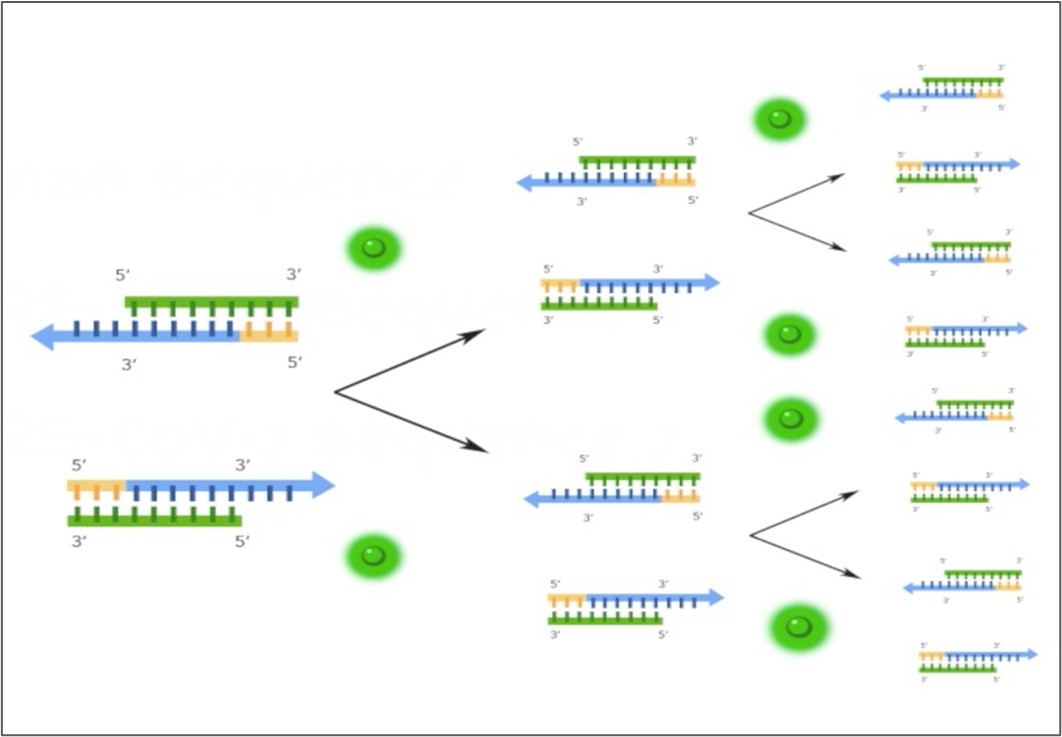
qPCR results in exponential amplification of a particular gene sequence, making it easy to detect with the use of sequence-specific fluorescent probes, represented by the green halos in the schematic.
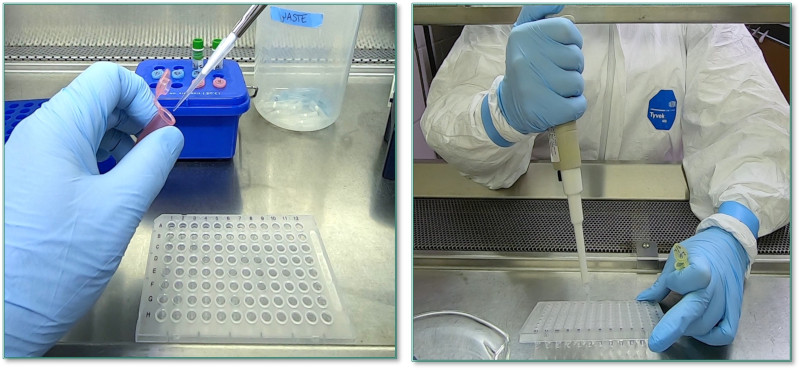
Each primer is mixed separately with the rest of the qPCR reagents and added to a 96-well plate (left). Samples of purified RNA are then added to each primer mixture (right).
Once all of the components are mixed with the purified sample RNA in a 96-well plate, they each play a part in amplifying the specific genetic sequences of the virus through a series of heating and cooling steps performed by the qPCR instrument (15). For instance, reverse transcription takes place first at 25-50C. Next, the reaction mixtures are heated to 94C, causing denaturation (16). At this step, the newly created double-stranded DNA separates into single strands. The temperature is then dropped to 55C, at which point annealing of the primers occurs, allowing the primers and fluorescent probes to bind to the gene of interest (17). The DNA polymerase then uses the primers to begin to create new copies of the gene of interest, releasing and activating the fluorescent probes with each successful amplification.
The denaturation and annealing cycles repeat through at least 40 cycles to yield either a positive or negative result for each set of primers. This process of exponential amplification can result in over a billion copies of the original piece of target DNA by cycle 30-35, depending on the starting concentration of the virus in the sample, or the viral load (18) The cycle threshold (Ct) value indicates the number of cycles performed by the instrument before the intensity of the fluorescent signal crosses the threshold for detection. A lower Ct value indicates greater starting concentration of SARS-CoV-2 templates, and therefore fewer amplifications required to reach the threshold for detection (19).
Click here to read the first publication from our COVID-19 clinical study: https://www.medrxiv.org/content/10.1101/2020.07.25.20160812v1

After loading the 96-well plate with the sample and qPCR mixtures, it is placed in a qPCR instrument.
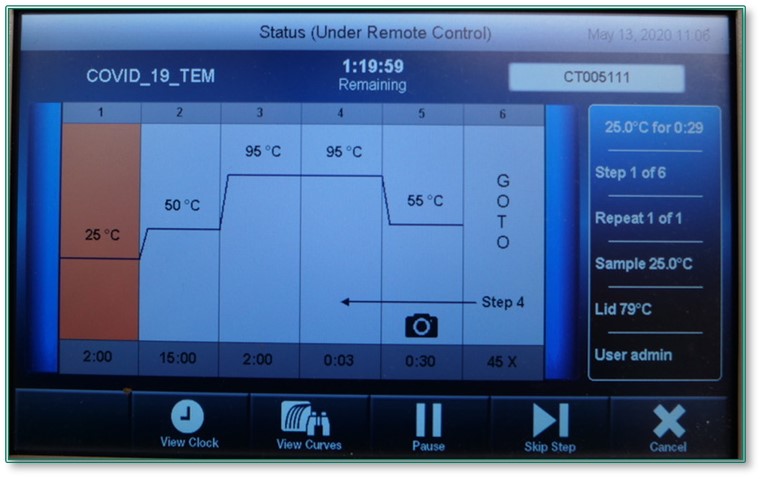
The protocol uses a one-step RT-qPCR assay, where the reverse transcription and polymerase chain reaction steps occur in the same tube.

To yield a positive result, amplification must reach the threshold of detection for both SARS-CoV-2 primers and the human gene primer. If the human sequence is not detected, the result is invalid. If only one virus primer is detected, the result is inconclusive. If all primers are detected, the result is positive.
References:
- https://en.wikipedia.org/wiki/RNA_extraction
- https://www.thermofisher.com/us/en/home/references/ambion-tech-support/rna-isolation/general-articles/the-basics-rna-isolation.html
- https://en.wikipedia.org/wiki/Spin_column-based_nucleic_acid_purification
- https://en.wikipedia.org/wiki/Lysis
- https://en.wikipedia.org/wiki/Elution
- https://en.wikipedia.org/wiki/Real-time_polymerase_chain_reaction
- https://www.nature.com/subjects/fluorescent-probes
- https://www.sigmaaldrich.com/life-science/custom-oligos/dna-probes/product-lines/fluorescent-probes.html
- https://www.sigmaaldrich.com/technical-documents/articles/biology/quantitative-pcr.html
- https://www.thermofisher.com/us/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/spotlight-articles/basic-principles-rt-qpcr.html
- https://www.biochain.com/general/using-dntp-in-polymerase-chain-reaction-pcr/
- https://www.nature.com/scitable/definition/primer-305/
- https://www.fda.gov/media/134922/download
- https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html
- https://www.medrxiv.org/content/10.1101/2020.07.25.20160812v1.full.pdf
- https://en.wikipedia.org/wiki/Denaturation_(biochemistry)
- https://en.wikipedia.org/wiki/Nucleic_acid_thermodynamics#Annealing
- https://www.promega.com/resources/guides/nucleic-acid-analysis/pcr-amplification/